
Received: FebruAccepted: JPublished: August 4, 2015Ĭopyright: © 2015 Hughes et al. Driscoll, The Francis Crick Institute, UNITED KINGDOM The decon1d program is freely available as a downloadable Python script at the project website ( ).Ĭitation: Hughes TS, Wilson HD, de Vera IMS, Kojetin DJ (2015) Deconvolution of Complex 1D NMR Spectra Using Objective Model Selection.
In contrast, the BIC method used by decond1d provides a quantitative method for model comparison that penalizes for model complexity helping to prevent over-fitting of the data and allows identification of the most parsimonious model. In current methods, determination of the deconvolution model best supported by the data is done manually through comparison of residual error values, which can be time consuming and requires model selection by the user.
#Mestrenova nmr peak broad software
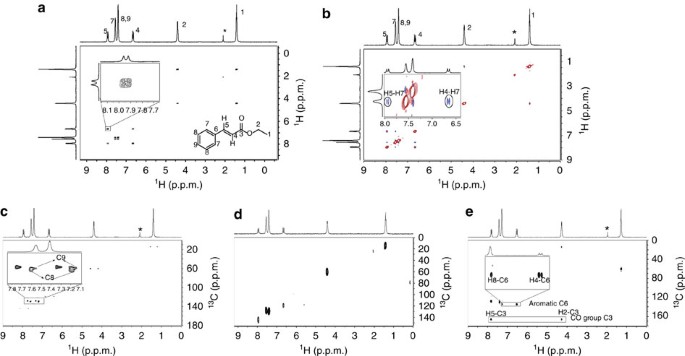
The method also allows for fitting of intermediate exchange spectra, which is not supported by current software in the absence of a specific kinetic model. We have developed a Python-based deconvolution program, decon1d, which uses Bayesian information criteria (BIC) to objectively determine which model (number of peaks) would most likely produce the experimentally obtained data. One-dimensional (1D) 19F NMR spectra of proteins can be broad, irregular and complex, due to exchange of probe nuclei between distinct electrostatic environments and therefore cannot be deconvoluted and analyzed in an objective way using currently available software. Post memes/jokes in /r/chemistrymemes and /r/chemistryjokes.Fluorine ( 19F) NMR has emerged as a useful tool for characterization of slow dynamics in 19F-labeled proteins. Any such posts will be deleted.Īsk education and jobs questions in the current weekly topic. If you're looking for a more concentrated, advanced discussion of chemistry topics among professionals and grad students, check out /r/Chempros.īefore asking "What chemical is this?" see this chart. Click here for the OSHA chemical data site and here for a multicompany MSDS aggregate search. If you spill/injure yourself contact medical professionals and read the MSDS, do not post to this reddit. Yes links to blogs, images, videos, comics, and infographics are okay especially if they are on your personal website.

No physorg, sciencedaily, or other press release aggregator spam!
#Mestrenova nmr peak broad license
If a caption or explanation is included this helps, but please use your discretion.īefore asking about chemical drawing/illustration programs, look at your school's IT/software website and see if they provide an institutional license of ChemDraw (hint: if they have a chemistry department, they will) Likewise, simple pictures of uninteresting and garden variety chemistry-related things are not appreciated. No memes, rage comics, image macros, reaction gifs, or other "zero-content" material. However, academic discussions on pharmaceutical chemistry and the science of explosives are permitted. Rules: Violating a rule will result in a ban.Īsk homework, exam, lab, and other undergraduate-level questions at ChemicalForums otherwise it will be deleted.ĭiscussions on illicit drug synthesis, bomb making, and other illegal activities are not allowed and will lead to a ban.


 0 kommentar(er)
0 kommentar(er)
